biomedical knowledge graph embeddings
Knowledge Graph Embeddings in the Biomedical Domain: Are They Useful? A Look at Link Prediction, Rule Learning, and Downstream Polypharmacy Tasks
This was a group project I worked on in the Master of Science by Research (MScR) degree part of the UKRI CDT in Biomedical AI at the University of Edinburgh. Aryo Pradipta Gema, Dominik Grabarczyk and I evaluated a bunch of knowledge graph embedding (KGE) models on a very large biomedical knowledge graph (KG). In the middle of the project, AstraZeneca published a paper that had the exact same goal for the exact same KG. A bit of a hit in the face to see your research idea scooped from under your nose, but our models seemed to be performing much better than theirs on the same data, so we pushed on a wrote an article about our work.
By embedding these massive KGs in lower dimensions, we can predict links that are missing in the KGs:
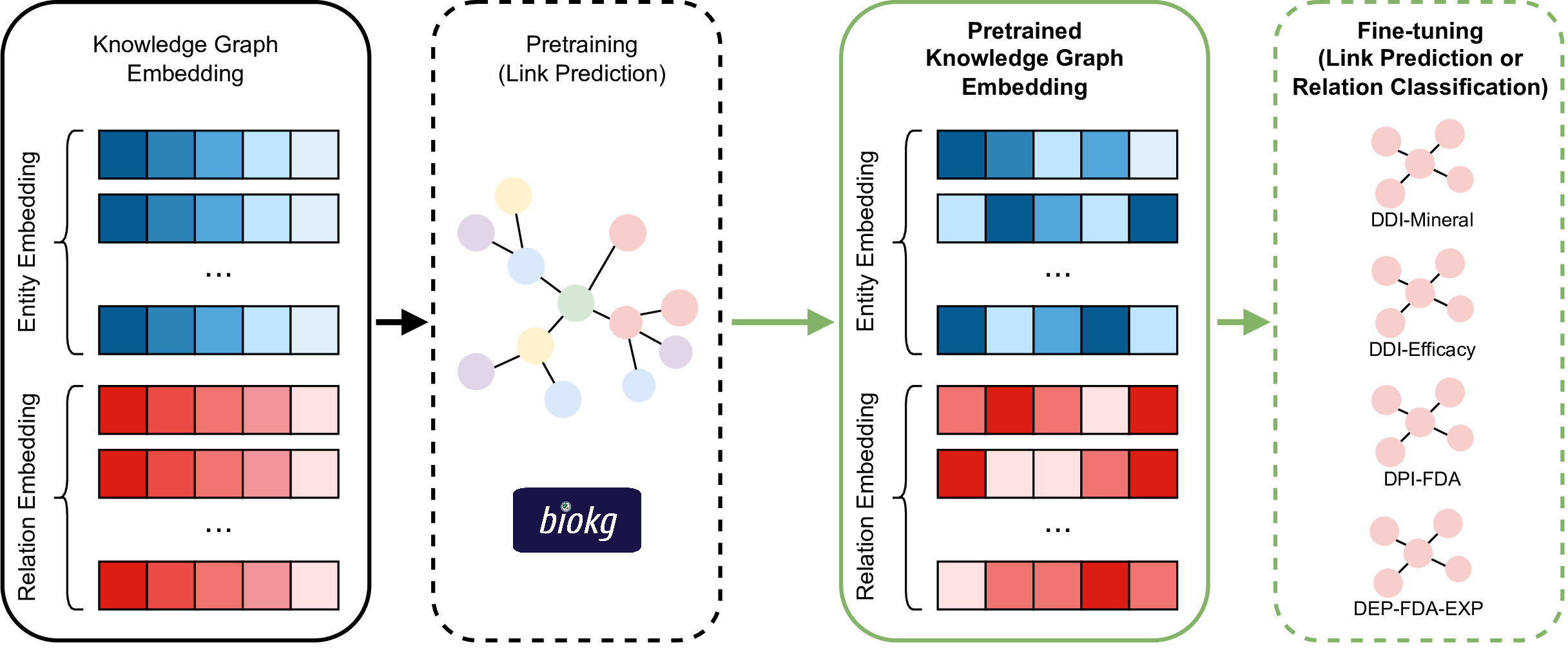
Depending on the content of the KG, new-found links can help clinical research such as drug repurposing or predicting side effects of drug combinations.
The article is out as a preprint on arXiv, and we’ve submitted it for publication in Bioinformatics Advances.
You can take a look at the GitHub repository here:
Supervisors: Dr. Ajitha Rajan, Dr. Pasquale Minervini, Dr. Antonio Vergari & Dr. Javier Alfaro